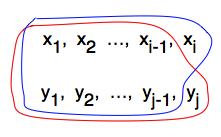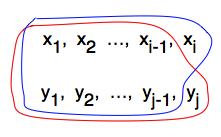SUNY Geneseo Department of Computer Science
Sequence Comparisons via Dynamic Programming
Thursday, March 14
CSci 242, Spring 2013
Prof. Doug Baldwin
Return to List of Lectures
Previous Lecture
Questions?
Problem 1 in problem set?
- Think of the choices Stacey has about what to do with the kth course
- She can take it, in which case she has ck more credits but pk fewer dollars to spend on the other courses
- Or she can not take it, in which case all her credits come from other courses, and all her money goes to take them
- She should make whichever of these choices leads to the most credits
- So the recurrence for her maximum number of credits for courses 1 through k and m dollars is
- maxCredits( k, m ) = max( ck + maxCredits(k-1,m-pk), maxCredits(k-1,m) )
- maxCredits( 0, m ) = 0
Sequence Matching by Dynamic Programming
Biological application: how similar are 2 DNA sequences or proteins
Longest common subsequence
- Section 15.4
- A G T matches A C C C C G C T and T A A G T C
- C(i,j) = length of longest common subsequence of X1..i and Y1..j
- = 0 if i = 0 or j = 0
- = C(i-1,j-1) + 1 if i, j > 0 and Xi = Yj
- = max( C(i-1,j), C(i,j-1) if i, j > 0 and Xi ≠ Yj
- Reflects two ways to find a longest common subsequence when you can’t automatically stick the last element of X and Y at the end
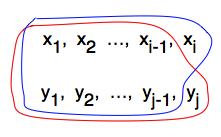
Needleman-Wunsch algorithm
- DNA sequences X = x1, x2, …, xn and Y = y1, y2, …, ym where xi, yi come from {A,C,G,T}
- Want to match X and Y so “similar” bases match, possibly by inserting gaps

- Similarity measured by function S(a,b) for a, b in {A,C,G,T} and gap penalty d
- Maximize sum over pairs in final alignment of S(a,b) where alignment pairs a and b and d where alignment inserts a gap in either sequence
- Element i of X and element j of Y can be paired at cost S(xi,yj), or either can be paired with a gap in the other sequence, leaving all i (or j) elements of that other sequence to align with the remaining elements in this sequence
- Recurrence for C(i,j) = maximum similarity score for aligning X1..i with Y1..j
- C(i,0) = d×i
- C(0,j) = d×j
- C(i,j) = max( S(Xi,Yj) + C(i-1,j-1), d + max( C(i-1,j), C(i,j-1) ) )
- Code for an iterative version
score( X, Y, n, m )
let C[0..n,0..m] be a new table
// C[i,j] = max score aligning X1..i w/ Y1..j
for i = 0 to n
C[i,0] = d * i
for j = 0 to m
C[0,j] = d * j
for i = 1 to n
for j = 1 to m
C[i,j] = max( S(Xi,Yj)+C[i-1,j-1], d + C[i-1,j], d + C[i,j-1] )
return C[n,m]
Next
Shortest paths in graphs
Section 25.2 through “Constructing a shortest path”
- Top of page 693 through first 1/3 of 697
Next Lecture